Scan Information File (SIF)
What is SIF data
SIF contains the PET frame start and end times, and the numbers of observed events during each PET time frame.
SIF data is stored in an ASCII text file. This is an example of a SIF data:
4/12/2002 18:11:32 28 4 1 B04946 C-11
0 60 125296 119001
60 75 637313 269790
75 80 1031361 299114
80 90 3196802 1001146
90 100 4401745 1349903
...
Title line contains the following information:
- Scan start time
- Number of frames
- Number of columns in SIF data
- SIF version (in this case 1)
- Study id (study number)
- Isotope that was used to label the PET radiopharmaceutical
Each data line contains the following columns:
- Frame start time (s)
- Frame end time (s)
- Number of prompts
- Number of random events
Why is SIF needed
Calculating fit weights for analysis models
Prompts and randoms in the SIF data can be used or are required in calculating weights for model fits. SIF data is required e.g. when applying simplified reference tissue model with basis function approach (imgbfbp). Weights can be added into regional datafiles using tacweigh.
Storing PET frame times with Analyze and NIfTI image formats
Analyze and NIfTI image formats do not contain information on PET frame times, therefore PET images in Analyze and NIfTI format should always be accompanied by SIF data.
DICOM images should contain frame times, but if not,
Carimas can read frame times from sif file saved in the DICOM folder with name
FrameInfo.sif.
With modern PET scanners, the prompt numbers are huge, which can be a problem for some software (such as Carimas 2.10). In that case you can set the prompts and randoms to zero in all SIFs in the current working folder in Windows command prompt with command:
for %g in (*.sif) do taccalc --force %g x 0 %g
Storing the label isotope with Analyze and NIfTI image formats
Analyze and NIfTI image format does not contain information on the isotope or its half-life, therefore dynamic Analyze and NIfTI images should always be accompanied by SIF data. If SIF does not either contain isotope, it can be added to SIF files using sifisot.
Where does SIF data come from
From PET scanners
PET image data is stored in PACS in DICOM format which contains the time frame information. If image data is converted to Analyze 7.5 or NIfTI format using ImageConverter, be sure to tick the check-box for saving SIF.
GE Advance
In Turku PET Centre, SIF data is extracted together with ECAT 6.3 images from GE Advance database, and written in the same directory as the PET image with extension .sif.
HR+
SIF data can be extracted from HR+ scan data (*.s) using hrp2sif.
Siemens Inveon small-animal PET/CT
SIF data can be extracted from original image header files (*.hdr) using upet2sif.
From dynamic PET image
If SIF data is not available, the weights can be estimated with sufficient accuracy based on PET image data. Programs eframe and imgweigh can be used to produce SIF from dynamic ECAT 6.3 or ECAT 7 images.
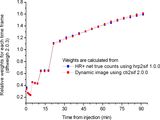
Comparison of fit weight factors calculated from net true counts (gold standard) and from average radioactivity concentration in the dynamic image:
Brain [18F]FDOPA study scanned with HR+:
Brain [11C]FLB study scanned with HR+:
Brain [11C]PIB study scanned with HR+:
Myocardial [11C]HED study scanned in 2D with HR+:
Myocardial [18F]FDG study scanned in 2D with HR+:
Myocardial [15O]H2O study scanned in 2D with HR+:
From regional PET curves
If absolutely necessary, the SIF files can also be made from regional PET data using
tacweigh with option -sif=filename.
Program tacframe can make SIF from
TTAC file containing just the frame times.
Software for processing SIF data
- Creating SIF data from ECAT image or sinogram, or specifically from dynamic HR+ sinogram
- For Viewing the SIF data use
taclist, preferably with option
-ift - Catenating SIF data from interrupted PET scans
- Adding isotope code to SIF.
- Setting study number to SIF.
See also:
Tags: SIF, Time frame, File format, Analyze 7.5, NIfTI, Weighting, Carimas
Updated at: 2020-02-28
Created at: 2013-05-14
Written by: Vesa Oikonen