MPI with chemical microspheres
Labelled small microspheres that are trapped in tissue capillaries can be used to quantify nutritive blood flow to tissue (perfusion). Similarly, perfusion could be assessed with labelled radiopharmaceutical that is rapidly and irreversibly taken up by cells.
While in this model configuration the arterial plasma curve is used as the input function, in the more practical model setting (Fig 2) the blood curve is used instead.
Concentration of the tracer in the blood (CB) is an average of concentrations in plasma (CP) and red blood cells (CRBC), weighted by haematocrit (HCT):
The arterial plasma TAC, not the whole blood TAC, is the correct input function in this model since the radiopharmaceutical in blood cells is assumed to be trapped. Since we want to assess blood flow instead of plasma flow, we write the differential equations for tissue and blood cell concentrations as
, where (1-HCT)×CP is the plasma radioactivity per volume of blood. After integration and rearrangement,
, which clearly shows that in this chemical microsphere model the TACs of muscle tissues and RBCs have the same shape, the integral of plasma TAC as a function of time, but with their own scaling factors.
Solving Eq 4 for CRBC(t) and substituting it into Eq 1 gives equation
which can be solved for (1-VB)×CP(t), which is then substituted in Eq 2, giving
This is similar to the differential equation of the commonly used reversible one-tissue compartment model with k2=kC×HCT.
The regional TACs from myocardial PET studies are distorted by Spillover and partial volume effects. In theory, this can be accounted for by modelling the left ventricular (LV) muscle and cavity TACs as:
The β is dependent on the image resolution, and could be measured with [15O]CO PET. In myocardial radiowater PET studies the model can be first fitted to the data from a large ROI covering most of the left ventricular myocardium (possibly excluding septum and areas with marked spillover from the liver), enabling calculation of CB(t), which is then used for analysis of smaller myocardial regions (Iida et al., 1988, 1991, and 1992). The resolution of modern PET may allow using the blood curve derived from a tiny ROI placed in the LV cavity directly, and therefore we assume that β=1, and
While the α can be estimated in myocardial PET studies with radiowater that is a diffusible tracer with known equilibrium tissue distribution volume, that is not possible with a trapped radiopharmaceutical. Instead we must draw muscle ROI avoiding carefully the tissues outside of the heart, and use the simple geometrical model
CT(t) and dCT(t)/dt are solved from this equation and its derivative, respectively, and placed in Eq 6, providing the ODE and its integral
This multilinear equation has three parameters, VB, K1, and k2=kC×HCT, and all concentrations are measured with PET. The parameters can be fitted with non-linear optimisation methods, or solved using non-negative least squares (NNLS) optimisation method, like has been done for example to fit muscle data from radiowater PET studies (Burchert et al., 1997). The NNLS method is computationally very fast, allowing pixel-by-pixel calculation and production of parametric K1 and VB image.
The last of the parameters, k2=kC×HCT, is related only to the kinetics of tracer in blood and plasma, and therefore common to all tissue regions. Therefore it is possible to either perform the fits of all tissue regions simultaneously, with a single k2 parameter, or to first fit a large (less noisy) myocardial region, and fit the (noisier) TACs of smaller myocardial regions with fixed k2. If k2 is known, the remaining two parameters can be solved using NNLS from multilinear equation
With k2, the plasma component of measured, discrete blood data curve could be computed from ODE that could, for example, be solved using implicit second-order Adams-Moulton corrector, resulting in equation
, where Δt is the sample time difference. Program b2ptrap can make the plasma curve with this method using known k2, or it can fit the k2 based on blood data alone with assumption that radioactivity in plasma is zero after a certain time. Program fitmtrap, which can be used to fit the model in Eq 10 to regional data, can also make the plasma curve with this method.
If both arterial plasma and blood curves would available, a very simple linear equation can be formed with only two parameters to be fitted:
The reversible one-tissue model with parameters VB, K1, and k2 is included in all PET modelling software, and the parameters VB and K1 from those tools can directly be interpreted as the parameters of the chemical microsphere model.
K1 and perfusion
The unidirectional transport rate of the radioligand from the blood to the tissue (K1) and blood flow (F), according to the Fick's principle and Renkin-Crone capillary model, are related by first-pass extraction fraction E,
, where E depends on perfusion, and the product of capillary permeability P and capillary surface area S, PS. Only highly diffusive (E ≅ 1) tracers can be used to reliably measure regional perfusion. If E is substantially reduced when perfusion is high, a non-linear function is needed to convert K1 to perfusion. E cannot be determined from PET data, but comparison of K1 estimates to perfusion values measured with gold-standard methods can provide the non-linear function that can be used to obtain perfusion from K1.
Simulations
Late-time tissue-to-blood ratio (TBR)
An intravenously administered radiotracer that traps into cells is rapidly cleared from the plasma. As plasma curve approaches zero, its area-under-curve (integral) stabilizes to a certain level. Likewise, the concentrations in blood cells and tissue stabilize to a level determined by the integral (area-under-curve) of the plasma concentration and the rate constants k2=kc×HCT and K1 (see Eq 4). At this phase, since CP=0, the concentration in blood is
and the late-time regional tissue-to-blood ratio, based on Eq 7 and CT/CRBC from Eq 4, is
We can see that the late-time TBR correlates with K1, but is affected by VB (which represents the partial volume and spillover effects in myocardial model) and k2=kc×HCT (the rate of tracer uptake in blood cells). The relationship of K1 and late-time TBR is shown in Fig 3. When K1 < kC×HCT, the late-time concentration in myocardial regions will be lower than concentration in blood (LV cavity), which complicates the drawing of regions-of-interest to areas with perfusion defect.
Haematocrit has a linear effect on late-time TBR (Eq 13). The tissue uptake is related to plasma flow, and its proportion of blood flow, (1-HCT)×F, is higher when haematocrit is lower, which explains the higher late-time TBR with lower haematocrit in the right panel of Fig 3.
The late-time TBR correlates linearly with K1 when other model parameters are assumed constant. VB however has strong effect on the regional tissue concentrations. When drawing the regions on the PET image it is difficult to control the contribution of blood in LV cavity to the muscle regions; usage of tissue-to-blood ratio (TBR) or late-time SUV in quantification of myocardial perfusion cannot therefore be recommended, especially as the compartment model solutions are readily available, providing K1 that is not affected by VB, kC, or haematocrit. However, the late SUV or TBR image may be useful in visual analyses.
Kinetic data simulation
In this simulation, a sum of exponentials function (Feng et al., 1993a and 1993b) is used to describe the input function of the model, the arterial plasma curve (CP(t)), shown in Fig 4. In the present model the intravenously administered radiotracer is assumed to become rapidly trapped in cells, including blood cells, and therefore the function is set to approach zero in few minutes. The tracer concentration in blood cells is simulated based on Eq 3, and the arterial blood curve is calculated as the sum of tracer concentrations in blood cells and plasma (Fig 4).
Myocardial muscle tissue curves (CT) were then simulated using Eq 2 with K1 values ranging from 0.2 to 3.0 mL Blood/(min × mL tissue). These, and previously simulated blood curve, were used to calculate regional muscle curves (CPET) with different values for VB (Fig 5).
The simulated TACs reach a steady level ∼5 min after tracer administration, but the exact time depends on the input function and model parameters. As expected, the simulated, noiseless, regional tissue data could be well fitted with reversible one-tissue compartment model using blood curve as input function, and it provided K1 and VB estimates that were close to the ones used to simulate the data (results not shown). Extending the PET scan long after the steady phase is achieved would not help the parameter estimation; in real-world PET studies it may be detrimental, if radioactive metabolites of the tracer are released from cells at later times, invalidating the model.
PET image simulation
As a rudimentary mock-up of a myocardial dynamic PET image was made by filling segments of a ring with myocardial muscle TACs simulated with different K1 values, and centre with blood TAC, representing LV cavity (Fig 6 and Fig 7). The input curves simulated previously were used.
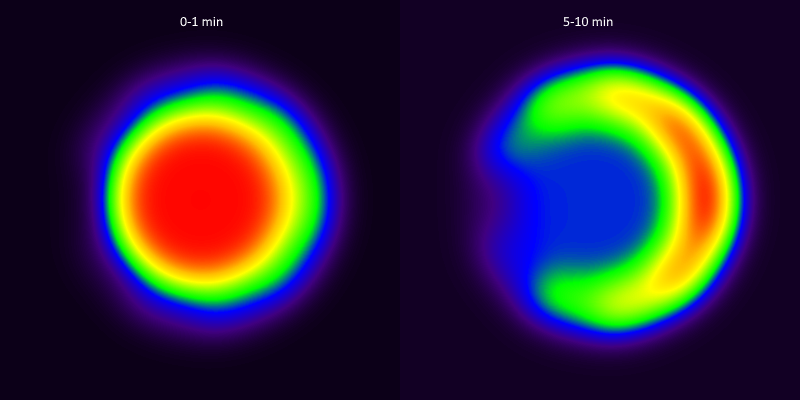
Dynamic image was then constructed with these TACs. In this simulation, no noise was added. Partial volume and spillover effects were simulated by strong smoothing with Gaussian filter; Fig 7 shows the sum images from time range 0-1 min and 5-10 min.
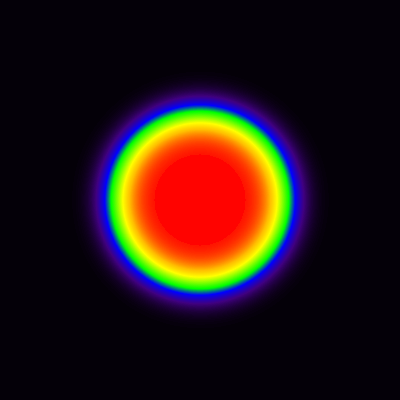
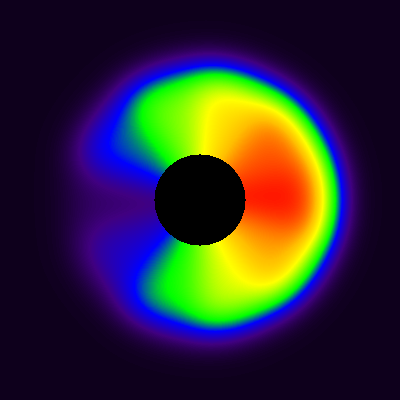
The blood curve from then extracted from middle of the area representing cavity, and model in Eq 9 was applied pixel-by-pixel to the dynamic image, producing parametric VB and K1 images (Fig 8). The spillover leads to apparent presence of muscle tissue far inside the cavity, in such extent that K1 can be estimated there, but not in the middle of the cavity where VB approaches 1, and K1 can no longer be estimated (represented by the black hole in the middle of the image where K1 is set to zero). Partial volume effect leads to underestimation of K1 in the region where the muscle actually is located, but less so at the border of muscle and cavity, because the spillover between cavity and muscle is corrected in the model (with the model parameter VB).
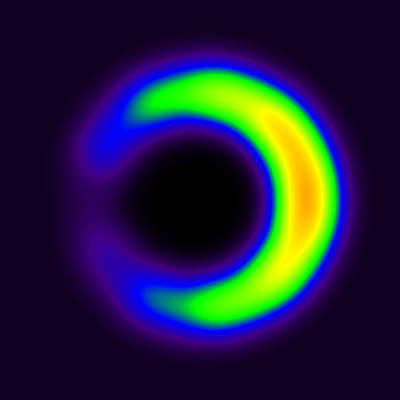
The effect of spillover on LV cavity
In the present model and previous simulations the assumption is that true arterial blood curve can be extracted from a small ROI drawn in the middle of the LV cavity. In practise, the blood curve may be contaminated by myocardial muscle curve because of spillover and partial volume effects. In this simulation, the cavity curves were simulated with β values 100%, 95%, 90%, and 85%, meaning contamination of 0%, 5%, 10%, and 15% from myocardium, respectively. Since the level of tissue curve is dependent on perfusion, the simulation is performed with three different K1 values, 0.5, 1.0, and 2.0. The simulated data were then fitted using fitmtrap according to Eq 9. The model fitted the simulated curves perfectly (not shown). The simulated cavity curves and myocardial curve, and the bias induced to the estimated model parameters, are shown in Fig 10. The simulations suggest that tissue-contaminated blood curve leads to overestimation of all model parameters. The bias in both K1 and VB is similar to the contamination-% of the cavity curve. While the higher perfusion leads to higher overestimation of cavity curve, this does not influence the bias in estimated K1 values, which seems to be overestimated by about the same percentage in all perfusion levels. Most of the bias seems to be absorbed by the parameter k2.
Overall, the results of this simulation suggest that the possible contamination of LV cavity curve by myocardial muscle causes overestimation of K1, the bias remains at acceptable level. On the other hand, the nuisance parameter k2 is severely biased, suggesting that k2 should not be constrained if contamination of LV cavity curve is suspected.
Script and data used to make these simulations are available in
https://gitlab.utu.fi/vesoik/simulations/.
See also:
Tags: Perfusion
Updated at: 2023-03-18
Created at: 2023-02-27
Written by: Vesa Oikonen