Hill function in plasma metabolite correction
Hill type functions have been often used to fit the fractions of parent radiotracer in plasma.
An extended Hill function can be used when the parent tracer fraction, fp, is initially 1 or less then 1 (in some cases radiopharmaceutical is metabolized already in vasculature before reaching sampling site), and then approaches 0 or a higher fraction:
, where 0 ≤ a ≤ d, b ≥ 1, c > 0, 0 < d ≤ 1, and e ≥ 0.
Parameter d represents the initial level of parent fraction, and parameter a the final level. Parameter e is the time after which the fraction starts to decrease (Tarkia et al., 2012).
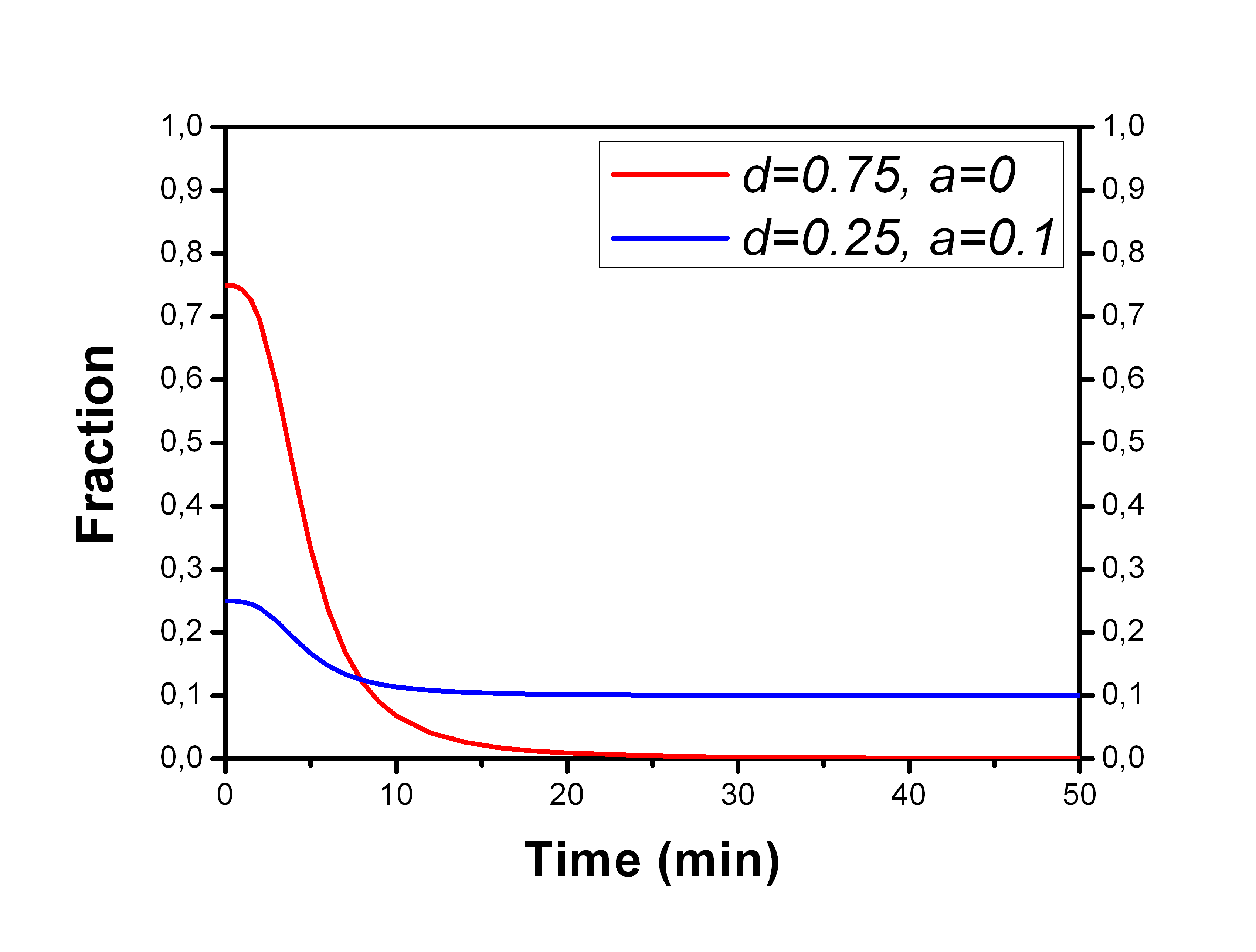
Unchanged (parent) tracer fraction curves can be fitted with fit_ppf with option -model=HILL.
If parameter d is set to 1 and e to 0, we have the traditional Hill type equation, which always starts from 1, but can still stop decreasing at a certain level (1-a) higher than 0:
Time when half of the isotope label carrying compounds in plasma are metabolites (t½) can then be calculated from equation:
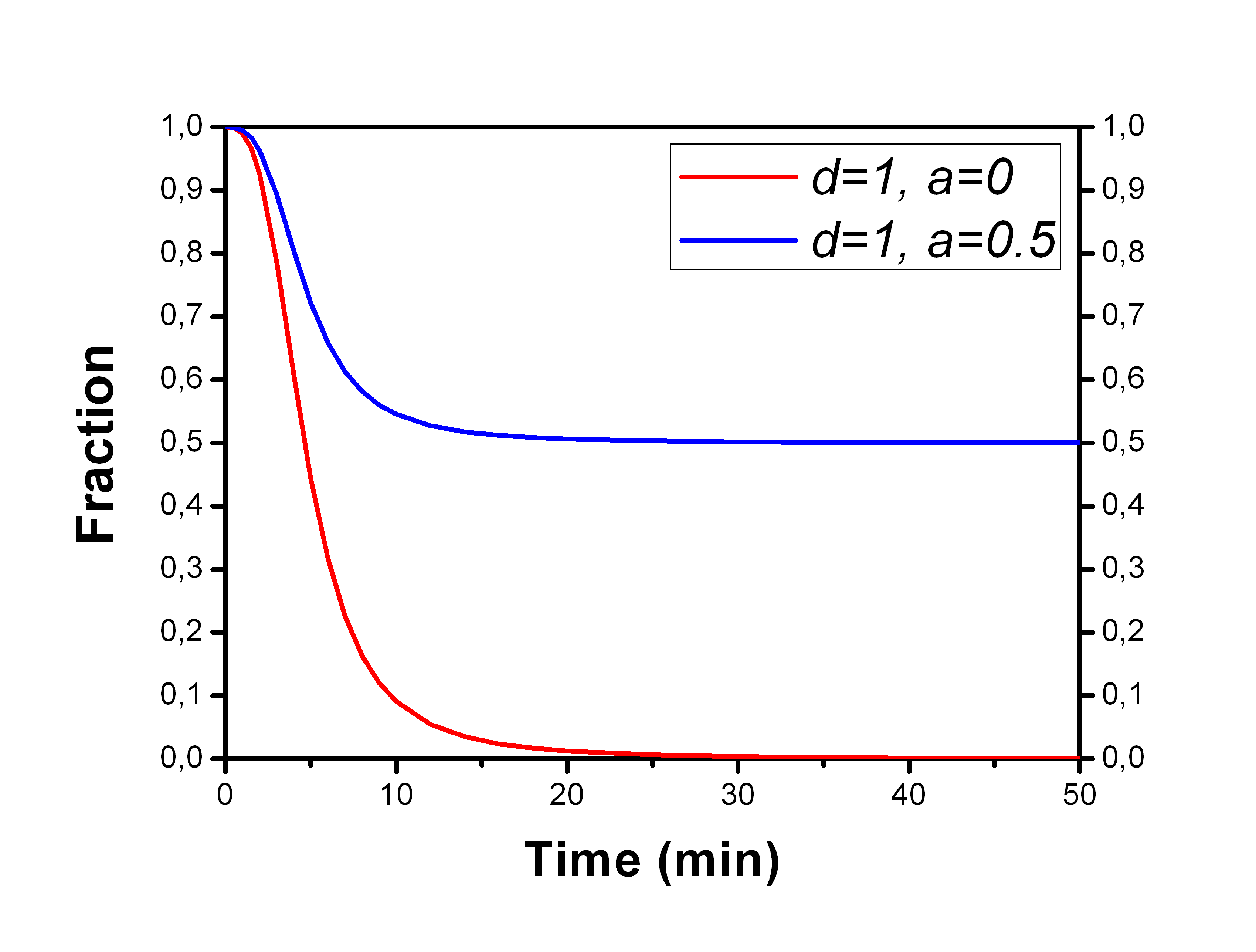
Both parent fractions and two metabolite fractions can be fitted simultaneously with fith2met, assuming that the a single Hill function shape can fit all the fractions.
Jucaite et al (2015) used a modified Hill function, mixture of the Hill and Richards equations (Giraldo, 2003; Bindslev, 2008). Guo et al (2013) used another modification:
If measured parent radiotracer fractions do not seem to reach a steady level during the measurement, or extrapolation is needed and you are not comfortable with the assumption of steady final level, then the power function should be applied instead.
The x axis value t for a known fp can be solved from the extended Hill function, giving equation:
The derivative of the extended Hill function is
See also:
- Power function in metabolite correction
- Metabolite correction
- Fractions of unchanged radiotracer in plasma
- Fitting PET input curves
References:
Bindslev N. Hill in hell. In: Drug-acceptor interactions. Co-Action Publishing; 2008. p. 257–82.
Giraldo J. Empirical models and Hill coefficients. Trend Pharmacol Sci. 2003; 24(2): 63-65. doi: 10.1016/S0165-6147(02)00048-2.
Guo Q, Colasanti A, Owen DR, Onega M, Kamalakaran A, Bennacef I, Matthews PM, Rabiner EA, Turkheimer FE, Gunn RN. Quantification of the specific translocator protein signal of 18F-PBR111 in healthy humans: a genetic polymorphism effect on in vivo binding. J Nucl Med. 2013; 54: 1915-1923. doi: 10.2967/jnumed.113.121020.
Jucaite A, Svenningsson P, Rinne JO, Cselényi Z, Varnäs K, Johnström P, Amini N, Kirjavainen A, Helin S, Minkwitz M, Kugler AR, Posener JA, Budd S, Halldin C, Varrone A, Farde L. Effect of the myeloperoxidase inhibitor AZD3241 on microglia: a PET study in Parkinson's disease. Brain 2015; 138(Pt 9): 2687-2700. doi: 10.1093/brain/awv184.
Wu S, Ogden RT, Mann JJ, Parsey RV. Optimal metabolite curve fitting for kinetic modeling of 11C-WAY-100635. J Nucl Med. 2007; 48: 926-931. doi: 10.2967/jnumed.106.038075.
Tags: Metabolite correction, Parent fraction, Fitting
Updated at: 2020-02-11
Created at: 2007-07-18
Written by: Vesa Oikonen